Finding simple systems which spontaneously generate self-replicating groups of cells from a random soup seems to have implications for the Origin of Life on Earth. This post explores those implications.
Theories on the Origin of Life
Wikipedia has a good summary on abiogenesis, or the Origin of Life, and talkorigins.org also has a good summary. One of the prevalent theories is the RNA World where the first self-replicating forms were RNA molecules that could catalyze the formation of more copies of themselves. This idea is conceptually simple, but it would be extremely rare that one of these molecules could form spontaneously (fortunately, life had millions--and perhaps billions--of years to originate).
But what if the first cells had a collection of simpler molecules that in combination could replicate themselves. This set of molecules is known as an Autocatalytic Set. And this is how modern cells replicate themselves, although the current system contains roughly a million molecules and includes DNA as the library from which all other molecules are made. Even a simpler autocatalytic set of a few dozen molecules would necessarily involve a complex process with many interactions. Aren't the chances of finding an autocatalytic set of simpler molecules just as unlikely as finding a self-replicating RNA molecule? Perhaps not! The emergent self-replicating systems presented on this website may provide support for the Autocatalytic-Set hypothesis.
Cellular Automata and Autocatalytic Sets
Consider this analogy:
- Each cell (either filled for empty) in the grid is a molecule
- Each unique cell state (empty/filled & number of neighbors) corresponds to a different type of molecule (even though they are represented by only two colors)
- Each iteration in the cellular automata corresponds to the molecules doing a reaction whereby they can change into other molecules (i.e. change state)
When we run one of the cellular automata systems with emergent self-replication, a pattern of cells ends up replicating itself, although not every type of cell is present at all times. This corresponds to a set of molecules replicating itself, even though all types of molecules are not present at the same time.
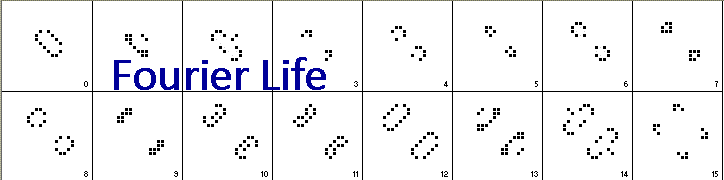
Let's consider an example. At iteration 3 below (the fourth frame), there are two patterns each with the following types of filled cells (n=neighbors): 1x1n, 2x2n, 1x3n and the following empty cells (excluding ones with 0 neighbors): 8x1n, 6x2n, 1x4n. But at iteration 4, there are two patterns with these filled cells: 1x0n, 2x1n, 3x2n and these empty cells: 12x1n, 9x2n, 2x3n, 1x4n. The set of cell types has changed dramatically in a seemingly non-productive way. However, the cell types keep changing until we get to iteration 15 where the group of cells at iteration 3 has replicated itself! Thus, the exact number of the different cell types at iteration 3 had doubled (at the expense of the empty 0-neighbor cells), and this process continues with each replication period. Throughout the process, the "autocatalytic set" of cell types replicates itself, but in a messy process involving many intermediate steps, and it is difficult to see the process when viewed at the cell-type level. This is very similar to what we see with molecules in living cells today.
Let's consider an example. At iteration 3 below (the fourth frame), there are two patterns each with the following types of filled cells (n=neighbors): 1x1n, 2x2n, 1x3n and the following empty cells (excluding ones with 0 neighbors): 8x1n, 6x2n, 1x4n. But at iteration 4, there are two patterns with these filled cells: 1x0n, 2x1n, 3x2n and these empty cells: 12x1n, 9x2n, 2x3n, 1x4n. The set of cell types has changed dramatically in a seemingly non-productive way. However, the cell types keep changing until we get to iteration 15 where the group of cells at iteration 3 has replicated itself! Thus, the exact number of the different cell types at iteration 3 had doubled (at the expense of the empty 0-neighbor cells), and this process continues with each replication period. Throughout the process, the "autocatalytic set" of cell types replicates itself, but in a messy process involving many intermediate steps, and it is difficult to see the process when viewed at the cell-type level. This is very similar to what we see with molecules in living cells today.
More than a hundred different emergent self-replicating rule-sets have been found in simple cellular automata systems using the Fourier Transform method. If the analogy above is correct, the soup of actual prebiotic molecules may also have many possible combinations which create autocatalytic sets. Perhaps the Fourier Transform method can be applied to finding such sets in computer simulations.
My Favorite Theory on the Origin of Life
I like the paper by Hordijk, Hein and Steel entitled Autocatalytic Sets and the Origin of Life. They present a convincing argument that life may have started with autocatalytic sets. They also point out that if the set was contained inside a liposome which would allow food substrates to enter, the first cell could have been born.
I'll add to the theory above that if the autocatalytic set also included the machinery to make the liposome lipids, the whole system could replicate itself. Perhaps the primordial soup contained catalytic molecules which could carry out various catalytic reactions, including the ones necessary to extend aliphatic carbon chains to make lipids. Once the lipid concentrations are high enough (either through synthesis or concentration by evaporation), liposomes would form with random combinations of molecules trapped inside. If just one of these new lipsomes contained an autocatalytic set--including the ability to make more lipid molecules--the cell would continue to grow. New lipid molecules would become integrated into the liposome and when it became big enough, it would automatically divide. Each new liposome would (in most cases) contain the autocatalytic set of molecules and the process would start again. If this is indeed the process which occurred, this would then be the moment when life began.
I'll add to the theory above that if the autocatalytic set also included the machinery to make the liposome lipids, the whole system could replicate itself. Perhaps the primordial soup contained catalytic molecules which could carry out various catalytic reactions, including the ones necessary to extend aliphatic carbon chains to make lipids. Once the lipid concentrations are high enough (either through synthesis or concentration by evaporation), liposomes would form with random combinations of molecules trapped inside. If just one of these new lipsomes contained an autocatalytic set--including the ability to make more lipid molecules--the cell would continue to grow. New lipid molecules would become integrated into the liposome and when it became big enough, it would automatically divide. Each new liposome would (in most cases) contain the autocatalytic set of molecules and the process would start again. If this is indeed the process which occurred, this would then be the moment when life began.